3 D Models
This is a description of the Java-free 3D viewer.
In MGDCOM more than 8,700 structural datasets of polyatomic molecules can be visualized as colored ball-stick models.
The access to the 3D-viewer can be recognized by a blue hyperlink in the single document window at the end of identification data below the structural formula and, if available, the graphic definition of the internuclear distances, bond angles and dihedral angles. The hyperlinks are always called "Show three dimensional molecular structure".
Then a new window is opened and a ball-stick model is displayed in the principal axis system of inertia. By clicking the axes icon [15] the principal axes will disappear; by another click on the icon the principal axes will appear again.
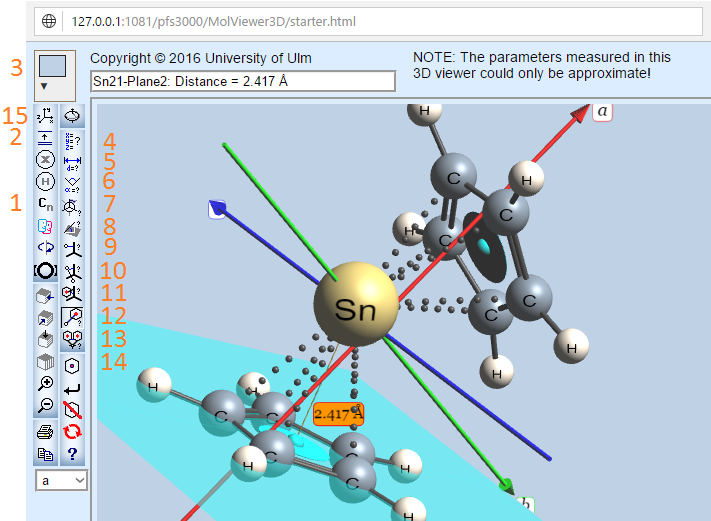
Additionally to the icons the displayed 3D-models can be interactively rotated, shifted, and scaled also by mouse movements:
rotate molecule | press left mouse button + mouse mouse |
shift molecule | press right mouse button + mouse mouse |
continous zoom-in | press middle mouse button + move mouse downward |
continous zoom-out | press middle mouse button + move mouse upward |
Further important icons:
- Show or hide atom labels
- Show or ignore bond order
- Select background color
- Display atomic Cartesian coordinates
- Calculate distance between atoms (select 2 atoms/centroids)
- Calculate angle between bonds (select 3 atoms/centroids)
- Calculate dihedral angle (select 4 atoms/centroids)
- Calculate elevation angle (select 4 atoms/centroids)
- Calculate distance between atom and axis (select 1 atom/centroid)
- Calculate angle between bond and axis (select 2 atoms/centroids)
- Calculate angle between axis and plane (select 1 centroid)
- Calculate distance between atom and plane (select 1 atom and 1 centroid)
- Calculate angle between two planes (select 2 centroid)
- Define centroid and least-squares plane (select at least 3 atoms and press Enter)
To calculate a geometrical property you have to click on the corresponding icon and then on up to 4 atoms/centroids.
